The incredible diversity of B and T lymphocyte receptors is the key to the adaptive immune system [1]. For high-throughput sequencing of the variable domains of these antibodies, total RNA is extracted and reverse transcribed using proprietary gene-specific primers that bind to the constant regions (Figure 1.1). The resulting cDNA is amplified using proprietary gene-specific primers to create next-generation sequencing libraries. These libraries were then subjected to deep sequencing.
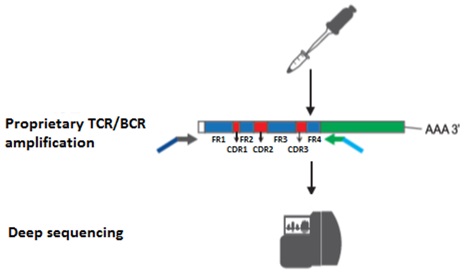
Figure 1.1 Immune repertoire sequencing (IgSeq) workflow
The raw sequencing data are in fastq format and can be found in FASTQ folder.The raw data were subjected to quality filtering. Sequences that passed the quality filtering were mapped against either IMGT database [2] or customized databases to find the best germline V(D)J gene matches. CDR sequences were then further characterized and analyzed.
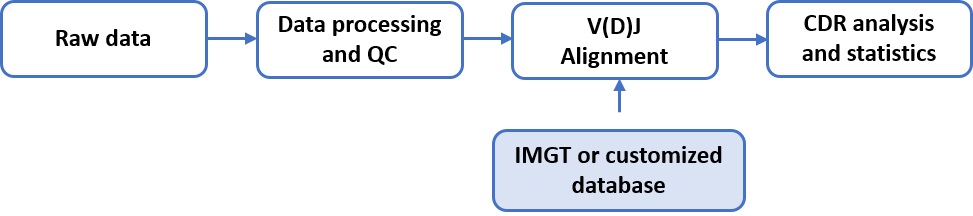
Figure 2.1 Bioinformatics analysis workflow
Raw fastq files were first subject to quality assessment [4] (Figure 3.1.1). Bases with poor quality scores (Q<20) were removed using Trimmomatic (v0.30) [3]. After quality trimming, reads shorter than 150bp were also removed from subsequent processing. Trimmed data were also subject to quality assessment [4] (Figure 3.1.2).
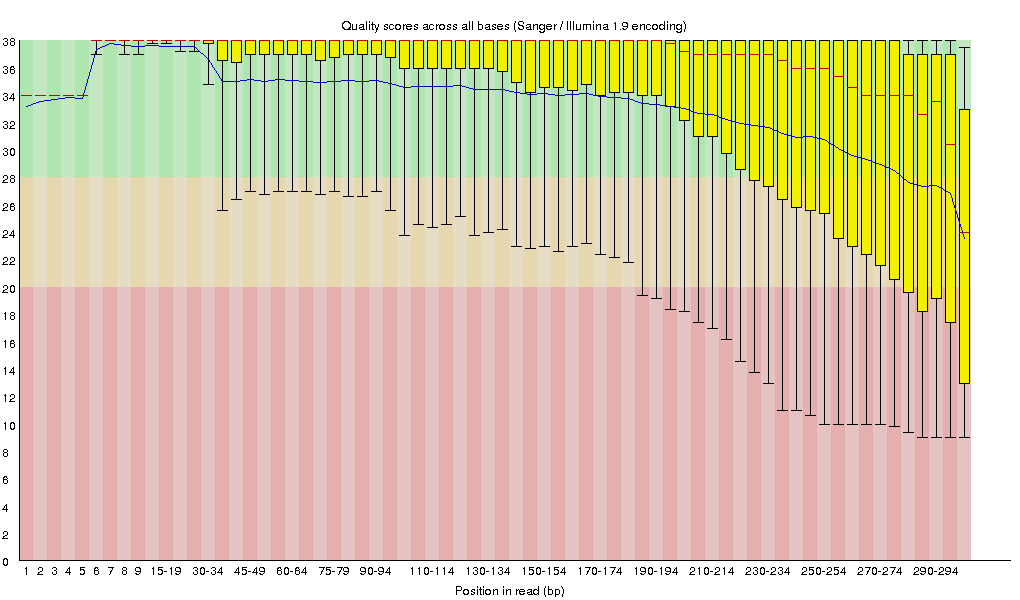
Figure 3.1.1 Sequence quality across all bases on raw reads.
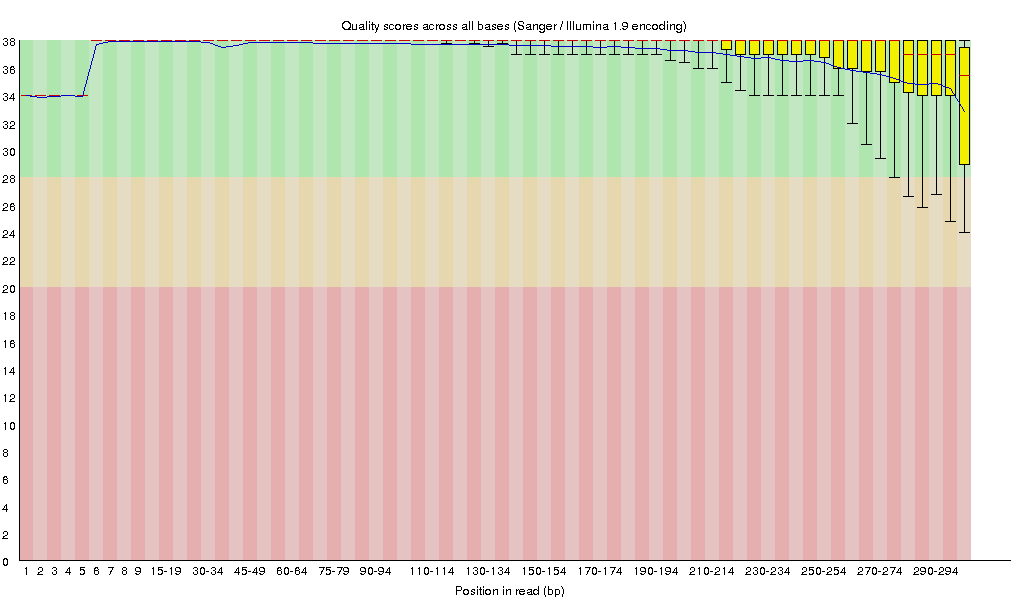
Figure 3.1.2 Sequence quality across all bases on clean reads.
After quality trimming, each Read1 and Read2 sequence pair was merged based on overlapping sequences. Merged reads with length less than 200bp were removed from subsequent analysis. The statistics of each processing step are summarized in Table 3.1. Length distribution of the merged contigs is shown in Figure 3.2.
Table 3.1 Raw sequencing data quality statistics
| Sample | Raw Count | % of Raw(>Q20) | % of Raw(>Q30) | Clean Count | % of Clean(>Q20) | % of Clean(>Q30) | % of Trimmed | Merged Count | % of Merged | Mapped Count | % of Mapped |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Raw_count | %_of_rawQ20 | %_of_rawQ30 | Clean_count | %_of_cleanQ20 | %_of_cleanQ30 | %_of_trimmed | Merged_count | %_of_merged | Mapped_count | %_of_mapped |
| ASC-S-vH | 1e+06 | 83.31 | 70.39 | 420350 | 99.08 | 94.94 | 42.03 | 300554 | 30.06 | 172313 | 57.33 |
| ASC-S-vL | 1e+06 | 85.27 | 73.77 | 447107 | 99.05 | 94.96 | 44.71 | 333677 | 33.37 | 119172 | 35.71 |
| PBMC-0-S-vH | 1e+06 | 83.78 | 71.34 | 448438 | 99.09 | 94.97 | 44.84 | 283774 | 28.38 | 173686 | 61.21 |
| PBMC-0-S-vL | 1e+06 | 85.08 | 73.35 | 431477 | 99.03 | 94.91 | 43.15 | 319008 | 31.9 | 114945 | 36.03 |
| PBMC-7-S-vH | 1e+06 | 83.71 | 71 | 439184 | 99.09 | 94.97 | 43.92 | 297230 | 29.72 | 171697 | 57.77 |
| PBMC-7-S-vL | 1e+06 | 85.15 | 73.38 | 427074 | 99.03 | 94.93 | 42.71 | 320174 | 32.02 | 116052 | 36.25 |
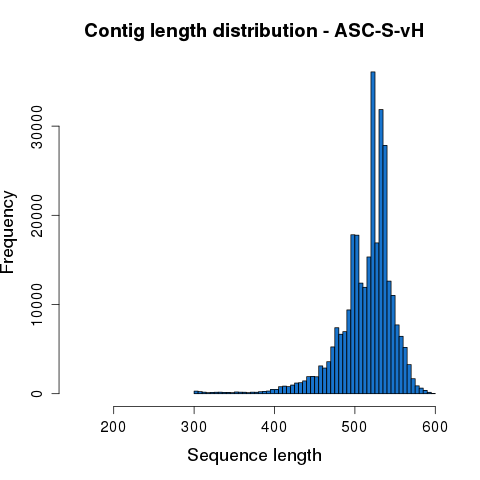
Figure 3.2 Assembled sequence length distribution.
The assembled reads were blasted against IMGT database to identify the best germline V(D)J gene matches [5]. The alignment result for each individual sequences is as in Table 4.1. The complete output for all the samples are in the ‘Report\Clonal_tables’ directory and can be accessed using the link below:
Table 4.1.1 VDJ gene mapping of clonal reads (heavy chain)
| Read_name | CDR1nt | CDR1aa | CDR2nt | CDR2aa | CDR3nt | CDR3aa | Top_V_Gene | Top_D_Gene | Top_J_Gene |
|---|---|---|---|---|---|---|---|---|---|
| M04670:18:000000000-ATLPC:1:2106:8878:1748:ACTCGCTA+CCTAGAGT | GGTGACTCCATCAGCAGTATTAGTTACTAC | GDSISSISYY | ATCTATTATAGTGGGAACACC | IYYSGNT | TGTGCGAGACATGGGGCGAGTAGTGCTAGTTGTTACCACGACTATTGG | CARHGASSASCYHDYW | IGHV3-5*02 | IGHD6-2*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:19468:2597:ACTCGCTA+CCTAGAGT | GGTGCCTCCGTCAGTAGTTACTAT | GASVSSYY | ATCTCTTTCAGTGGGATCACC | ISFSGIT | TGTGCGAGCCCCTGTTGGTATGACTGGAAGTGTCTCATGGACGTCTGG | CASPCWYDWKCLMDVW | IGHV3-2*02 | IGHD1-1*02 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:6990:3052:ACTCGCTA+CCTAGAGT | GGATTCACCTTCAGTAGCTATGCT | GFTFSSYA | ATATCATATGATGGAAGTAATAAA | ISYDGSNK | TGTGCGAGAGGTTTGGATCTGGGGGGCTACTACTACGGTATGGACGTCTGG | CARGLDLGGYYYGMDVW | IGHV5-17*02 | IGHD1-1*01 | IGHJ1*01 |
| M04670:18:000000000-ATLPC:1:2106:8601:2756:ACTCGCTA+CCTAGAGT | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATCAGGAGATCCGGGCTCCATAATGATGCTTTTGATATATGG | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHD2-3*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:6561:2747:ACTAGCTA+CCTAGAGT | NNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXX | NNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXX | TGTGTTAGAAGGCACCCAGCACCAACTGGCAACATTTTTGACTTCTGG | CVRRHPAPTGNIFDFW | IGHV5-9*02 | IGHD4-1*02 | IGHJ2*01 |
| M04670:18:000000000-ATLPC:1:2106:12327:1762:ACTCGCTA+CCTAGAGT | GGTGACTCCATCACCAGAGGTGATTACTTC | GDSITRGDYF | NNNNNNNNNNNNNNNNNNNNN | XXXXXXX | TGTGCGAGAGGTTCGGCCCCGACGGGGAACAACTACTTCGACCCCTGG | CARGSAPTGNNYFDPW | IGHV3-2*02 | IGHD2-1*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:15690:2814:ACTCGCTA+CCTAGAGT | GGGTTCTCACTCAACACTCGTGGAACGACT | GFSLNTRGTT | ATTTATTGGGATGGTGATGAC | IYWDGDD | TGTGCACACGGACGCCCAGACTGGGGAGCAGATGCTTTTGATGTCTGG | CAHGRPDWGADAFDVW | IGHV8-12*01 | IGHD4-1*01 | IGHJ1*01 |
| M04670:18:000000000-ATLPC:1:2106:23881:2561:ACTCGCTA+CCTAGAGT | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATCAGGAGATCCGGACTCCTTAATGATGCTTTTGATATCTGG | CARLPIRRSGLLNDAFDIW | IGHV3-2*02 | IGHD2-3*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:11036:3087:ACTCGCTA+CCTAGAGT | GGATTCTCACTGAGTCATAGTGGAGTGGGT | GFSLSHSGVG | ATTTATTGGGATGATGATAAA | IYWDDDK | TGTGCACACAGGGATGTAGTCACCTTTGACTCTTGG | CAHRDVVTFDSW | IGHV8-13*01 | IGHD1-1*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:17653:2689:ACTCGCTA+CCTAGAGT | GGTTACACCTTTACCAGCTATGGT | GYTFTSYG | ATCAGCGCTTACAATGGTAAGACA | ISAYNGKT | TGTGCGAGAGGGCAGTGGAACTATGATATAAGTGGAGCGTGGGACTACTGG | CARGQWNYDISGAWDYW | IGHV1S126*01 | IGHD2-4*01 | IGHJ4*01 |
| M04670:18:000000000-ATLPC:1:2106:15780:2769:ACTCGCTA+CCTAGAGT | NNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXX | NNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXX | TGTGCGAGAGATTACCACTGGTCTGGGTTTGACTACTGG | CARDYHWSGFDYW | IGHV5-17*01 | IGHD2-4*01 | IGHJ2*01 |
| M04670:18:000000000-ATLPC:1:2106:12947:3010:ACTCGCTA+CCTAGAGT | GGTGGGTCGTTTAGTGGTTTCGAC | GGSFSGFD | ATCAGCCACACTGGAACTACG | ISHTGTT | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV3-2*02 | IGHD2-3*01 | IGHJ3*02 |
| M04670:18:000000000-ATLPC:1:2106:13319:1512:ACTCGCTA+CCTAGAGT | NNNNNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXXX | NNNNNNNNNNNNNNNNNNNNN | XXXXXXX | TGTGCCCGTATAGTGGGAGGTAGTGTGGACTACTGG | CARIVGGSVDYW | IGHV3-1*02 | IGHD1-1*01 | IGHJ4*01 |
| M04670:18:000000000-ATLPC:1:2106:14336:3105:ACTCGCTA+CCTAGAGT | GATGGCTCCATCATCAGTGGTAATTACTAC | DGSIISGNYY | CGTATCTCT | RIS | TGTGCGAGATTTCTCGCTGGGAGTCAGTACCTTAACTACTGG | CARFLAGSQYLNYW | IGHV3-4*02 | IGHD5-1*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:11082:2663:ACTCGCTA+CCTAGAGT | GGTTACACCTTTACCAGATATGGT | GYTFTRYG | ACTAGCACTCACGATGAGGACTCA | TSTHDEDS | TGTGCGAGAGATTGGGACGGGAGAAACGACTGCTTCGACCCCTGG | CARDWDGRNDCFDPW | IGHV1-85*01 | IGHD4-1*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:16412:3064:ACTCGCTA+CCTAGAGT | GGTTACACCTTAACCACTTATGGT | GYTLTTYG | ATCGGCGCTTACAATGGTAACACA | IGAYNGNT | TGTGCGACAGACGGGCAGCAGCTGGTTCCGATGTCCGCCTGG | CATDGQQLVPMSAW | IGHV1-81*01 | IGHD5-7*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:14422:2879:ACTCGCTA+CCTAGAGT | GGTGCCTCCATCAGCACTAGTGATTACTAC | GASISTSDYY | ATCTATTATAGTGGGAGTAGC | IYYSGSS | TGTGCGAGACATTCTACAGATAGTACCGAGAAGTTCGACCCCTGG | CARHSTDSTEKFDPW | IGHV3-5*02 | IGHD2-5*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:10907:2923:ACTCGCTA+CCTAGAGT | GGGTTCTCACTCAACACTCCTGGAGTGTGT | GFSLNTPGVC | ATTGATTGGGATGATGATAAG | IDWDDDK | TGCGCACGGACGGATTGCAGTAACTACGGATGTTGGTTCGACCCCTGG | CARTDCSNYGCWFDPW | IGHV8-12*01 | IGHD2-5*01 | IGHJ3*01 |
| M04670:18:000000000-ATLPC:1:2106:16525:2101:ACTCGCTA+CCTAGAGT | GGGGACAGTGTCTCTAGCAACAGTGCTACT | GDSVSSNSAT | ACATACTACAGGTCCAAGTGGTATAAT | TYYRSKWYN | TGTACAAGAGGCTATAAGCAGCAAGACTACTGG | CTRGYKQQDYW | IGHV3S1*01 | IGHD3-2*02 | IGHJ4*01 |
| M04670:18:000000000-ATLPC:1:2106:20471:3646:ACTCGCTA+CCTAGAGT | GGTGCCTCCATTAACAGTGGTAGTTACTAT | GASINSGSYY | TTCTATACTACTGGGAGGACC | FYTTGRT | TGTGCGAGAGATCCCCCTCCCTACGTAATGGGAGCGCAGGCAGTGTGG | CARDPPPYVMGAQAVW | IGHV3-5*02 | IGHD5-7*01 | IGHJ1*01 |
Shown is the partial result of one sample (ASC-S-vH). Most of the sequences were truncated due to format limitation. Read_name: fastq header of the DNA sequence; CDR[1-3]nt/CDR[1-3]aa: the nucleotide/amino acid sequences of CDR1,2,3; Top_V_gene: the best matched germline v gene; Top_D_gene: the best matched germline D gene, Top_J_gene: the best matched germline J gene. Repetitive 'N's in nucleotide columns and repetitive 'X's in amino acid columns indicate no valid sequences were returned.
Table 4.1.2 VJ gene mapping of clonal reads (light chain)
| Read_name | CDR1nt | CDR1aa | CDR2nt | CDR2aa | CDR3nt | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|---|---|---|---|---|
| M04670:21:000000000-ATFAC:1:2106:21903:1604:GGAGCTAC+TCGACTAG | CAGGGCGTTGCCAGTGGT | QGVASG | GATGCCTCC | DAS | TGTCAGCAGTATAATAACTGGCCTTTGTGGACGTTC | CQQYNNWPLWTF | IGKV11-125*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:11438:1763:GGAGCTAC+TCGACTAG | CAGAGTGTTAGTAGCAAC | QSVSSN | GGCGCATCGAC | GAS | TGTCAGCAGTATGATAACTGGCCTTTGTGGACGTTC | CQQYDNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:10469:1752:GGAGCTAC+TCGACTAG | CAGACTATTAGCATCACCTAC | QTISITY | GGTGCATCC | GAS | TGTCAGCAGTACGGTAGTTCACCGTGGACGTTC | CQQYGSSPWTF | IGKV18-36*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:14873:1850:GGAGCTAC+TCGACTAG | NNNNNNNNNNNNNNNNNN | XXXXXX | NNNNNCTCC | XXS | TGTCAGCAATATTTTGTTACTCCGCTCACTTTC | CQQYFVTPLTF | IGKV18-36*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:16330:1899:GGAGCTAC+TCGACTAG | NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXXXXX | NNNNNNNNN | XXX | TGCATGCAATCTCTACAGACTCCTCTCACTTTC | CMQSLQTPLTF | IGKV2-a*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:13147:1941:GGAGCTAC+TCGACTAG | CAGACCATTACCAATTCC | QTITNS | GGTACATCC | GTS | TGTCAACAGAGTTACACCATCCCCTGGACGTTC | CQQSYTIPWTF | IGKV11-125*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:11304:1625:GGAGCTAC+TCGACTAG | NNNNNNNNNNNNNNNNNN | XXXXXX | GGTGCATCG | GAS | TGTCAGCAGTATAATAACTGGCCTTTGTGGACGTTC | CQQYNNWPLWTF | IGKV18-36*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:10681:1583:GGAGCTAC+TCGACTAG | CAGAGCATTAGCAGTGGT | QSISSG | GATGCCTCC | DAS | TGTCAACAATATAATAATTACCCCATCACCTTC | CQQYNNYPITF | IGKV11-125*01 | IGKJ4*01 |
| M04670:21:000000000-ATFAC:1:2106:17161:1570:GGAGCTAC+TCGACTAG | CAGAGTATTAGTACCCAG | QSISTQ | GAGGCATCT | EAS | TGCCTACAATATTTTTATTATTGGACGTTC | CLQYFYYWTF | IGKV11-125*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:15148:1742:GGAGCTAC+TCGACTAG | CAGAGTGTTAGCAGCTAC | QSVSSY | GATGCATCC | DAS | TGTCAGCAGTATAATAACTACTGGACGTTC | CQQYNNYWTF | IGKV18-36*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:12690:1861:GGAGCTAC+TCGACTAG | CAGAGCATTAGCGTCTAT | QSISVY | ACTGCATCC | TAS | TGTCAACAGAGTTACAGTCGCCCTCGGACGTTC | CQQSYSRPRTF | IGKV11-125*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:12369:1900:GGAGCTAC+TCGACTAG | NNNNNNNNNNNNNNNNNN | XXXXXX | NNNNNNNNN | XXX | TGTCAACAGCTTAGTAGTTACCCGCTCACTTTC | CQQLSSYPLTF | IGKV11-125*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:11496:1948:GGAGCTAC+TCGACTAG | AAGAGTGTTTTATCCATCTCCAACGACAAAAACTAC | KSVLSISNDKNY | TGGTCTTCT | WSS | TGTCAGCAGTATTATGATAAGCCGGTCACTTTC | CQQYYDKPVTF | IGKV8-30*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:13285:1814:GGAGCTAC+TCGACTAG | CAGGGCATAAGCAGGGGT | QGISRG | TATGCCTCC | YAS | TGTCAACAGTTTAATCAGTATCCCATCACGTTC | CQQFNQYPITF | IGKV11-125*01 | IGKJ4*01 |
| M04670:21:000000000-ATFAC:1:2106:16163:1988:GGAGCTAC+TCGACTAG | CAGGACATTGGCAGTTCT | QDIGSS | GGTGCATCC | GAS | TGTCAACAGCTTAAAAGTTACCCCATCAATTTC | CQQLKSYPINF | IGKV11-125*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:12925:2184:GGAGCTAC+TCGACTAG | CAGAGTGTTAGCAGCAAC | QSVSSN | GGTGCATCG | GAS | TGTCAGCAGTATAATAACTGGCCTTTGTGGACGTTC | CQQYNNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:13761:2193:GGAGCTAC+TCGACTAG | NNNNNNNNNNNNNNNNNN | XXXXXX | NNNNNNNNN | XXX | TGTCAGCAGTATGATAACTGGCCTTTGTGGACGTTC | CQQYDNWPLWTF | IGKV16-104*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:19380:2309:GGAGCTAC+TCGACTAG | CAGACTATTGGTGCCGACTAC | QTIGADY | GCTGCCTCC | AAS | TGTCAGCAGTATGGTACTTCACTTAGGACGTTC | CQQYGTSLRTF | IGKV18-36*01 | IGKJ1*01 |
| M04670:21:000000000-ATFAC:1:2106:19537:2312:GGAGCTAC+TCGACTAG | CAGAGCTTTGGCTGCTGC | QSFGCC | GGTGCCTCC | GAS | TGTCACCAGCGAAGTAGCTGGCCTCCGTTCACTTTC | CHQRSSWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| M04670:21:000000000-ATFAC:1:2106:24104:2288:CGAGCTAC+TCGACTAG | CAGAGTGTTCGCGGCAACTAC | QSVRGNY | GGTGC | G | TGTCAACAATATGGTAGCTCCACGTGGACGTTC | CQQYGSSTWTF | IGKV18-36*01 | IGKJ1*01 |
Shown is the partial result of one sample (ASC-S-vL). Most of the sequences were truncated due to format limitation.
To assess sequence abundancy, reads with the same CDR sequences were collapsed together and the counts of each unique sequences were calculated. The aggregation was performed on three levels – sequences were first collapsed based on nucleotide sequences of CDR1,2,3 (Tables 4.2.1 and 4.2.2), then further collapsed based on CDR1,2,3 amino acid sequences (Tables 4.2.3 and 4.2.4), and then further collapsed based on CDR3 amino acid sequence alone (Tables 4.2.5 and 4.2.6). The complete CDR1,2,3 enrichment analysis for all the samples are in “CDR_all_enrichment” directory, the CDR amino acid sequence enrichment analysis for all the samples are in “CDR_aa_enrichment” directory, and the CDR3 enrichment analysis for all the samples are in “CDR3_aa_enrichment” directory in the the Report/Enrichment_tables/ folder or can be accessed using the link below:
 CDR_all_enrichment
CDR_all_enrichment
 CDR_aa_enrichment
CDR_aa_enrichment
 CDR3_aa_enrichment
CDR3_aa_enrichment
Table 4.2.1 CDR enrichment by CDR1,2,3 nucleotide sequence (heavy chain)
| Count | CDR1nt | CDR1aa | CDR2nt | CDR2aa | CDR3nt | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|---|---|---|---|---|
| 11289 | GGTGGGTCGTTTAGTGGTTTCGAC | GGSFSGFD | ATCAGCCACACTGGAACTACG | ISHTGTT | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV3-2*02 | IGHJ3*02 |
| 3100 | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATCAGGAGATCCGGACTCCTTAATGATGCTTTTGATATCTGG | CARLPIRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 2339 | GGATTCATCTTCAGCACCTACTGG | GFIFSTYW | ATTAATGAAGATGGCAGGATTACC | INEDGRIT | TGTGTTAGAAGGCACCCAGCACCAACTGGCAACATTTTTGACTTCTGG | CVRRHPAPTGNIFDFW | IGHV5-6-3*01 | IGHJ2*01 |
| 1980 | GGTGACTCCATCAGCAATACTAGATATTAC | GDSISNTRYY | ATATATAATAGTGGAAATATC | IYNSGNI | TGTGCGGGGCACGTTTGGAACTACGAAGTTGACTACTGG | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 1452 | GGTGTCGCCATCACTAGTTTCCAC | GVAITSFH | ATATATCACAATGGAGACACC | IYHNGDT | TGTGCGAGAGTTGATGCAATCATTGAAATGGACTACTTCTACGGTCTGGACGTCTGG | CARVDAIIEMDYFYGLDVW | IGHV3-2*02 | IGHJ1*01 |
| 1339 | GGTGACTCCATCAGCAATACTAGATATTAC | GDSISNTRYY | ATATATAATAGTAGAAATATC | IYNSRNI | TGTGCGGGGCACGTTTGGAACTACGAAGTTGACTACTGG | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 976 | GGGTTCTCACTCAATACTCGTGGAACGACT | GFSLNTRGTT | ATTTATTGGGATGATGATAGC | IYWDDDS | TGTGCACACGGACGCCCAGACTGGGGAGCAGATGCTTTTGATGTCTGG | CAHGRPDWGADAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 931 | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATCAGGAGATCCGGGCTCCATAATGATGCTTTTGATATATGG | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 829 | GGCGGCTCCATCAGTAGCAGTAGTTACCAC | GGSISSSSYH | ATCTATTATAGTGGGAGCACG | IYYSGST | TGTGCGAGTCGTCGAAATGAACCTGGAGGGTGGTTCGACTCCTGG | CASRRNEPGGWFDSW | IGHV3-1*01 | IGHJ3*01 |
| 753 | GGTGGGTCATTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATGAGGAGATCCGGGCTCCTTAATGATGCTTTTGATATCTGG | CARLPMRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 705 | GGTTACACCTTTACTGATTATGCT | GYTFTDYA | ATCAGCGTTTCCAATGGTAAAACA | ISVSNGKT | TGTGCGAGAGCGTTTCAACCTCAAGTCTGGGTCGGGGAGTCTTATCTCGACTACTGG | CARAFQPQVWVGESYLDYW | IGHV1-77*01 | IGHJ2*01 |
| 666 | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTACC | ISHSGST | TGTGCGAGACTTCCCATCAGGAGATCCGGGCTCCATAATGATGCTTTTGATATATGG | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 626 | NNNNNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXXX | NNNNNNNNNNNNNNNNNNNNN | XXXXXXX | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV12-3*01 | IGHJ3*02 |
| 611 | GGGTTCTCATTCACTTCTAGTGGACGGGGT | GFSFTSSGRG | ATTTATTGGGATGACGATAAG | IYWDDDK | TGTGCACACAGACCACCATATCAAGGGTACTACTACTTTGACTATTGG | CAHRPPYQGYYYFDYW | IGHV8-12*01 | IGHJ2*01 |
| 602 | GGGTTCTCATTCAATACTCTTGGAACGACT | GFSFNTLGTT | ATTTATTGGGATGATGATAGC | IYWDDDS | TGTGCACACGGACGCCCAGACTGGGGAGAAGATGCTTTTGATGTCTGG | CAHGRPDWGEDAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 572 | GGCTTCTCATTCAGTTCTAGTGGACTGGGT | GFSFSSSGLG | ATTTATTGGGATGATGATAAG | IYWDDDK | TGTGCACACAGACCGCCATATCAACGGTATTACTACTTTGACTATTGG | CAHRPPYQRYYYFDYW | IGHV8-6*01 | IGHJ2*01 |
| 514 | GGATACACGTTCACCAATTATGCT | GYTFTNYA | ATGGACCCGAATAGCGGCGACACA | MDPNSGDT | TGTGCGAGGACCAACTGGGCAGCCTACGGTGTCCCCGACTACTGG | CARTNWAAYGVPDYW | IGHV1-72*04 | IGHJ4*01 |
| 485 | NNNNNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXXX | NNNNNNNNNNNNNNNNNNNNN | XXXXXXX | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV12-3*02 | IGHJ3*02 |
| 459 | GGTGACTCCGTCAGCAATCATAAATACTAC | GDSVSNHKYY | ATCTATTCTGGTGGGAACACC | IYSGGNT | TGTGCGGGGCACAATTGGAATTACGAGGTTGACTACTGG | CAGHNWNYEVDYW | IGHV3-5*02 | IGHJ4*01 |
| 455 | GGATTCTCACTCACTACTCGTGGAGGGGGT | GFSLTTRGGG | CTTTATTGGGATGAAAAGACA | LYWDEKT | TGTGCACACCGTATTGGTTCGGAGACATACTTCGACTACTGG | CAHRIGSETYFDYW | IGHV8-12*01 | IGHJ2*01 |
Shown is the partial result of one sample (ASC-S-vH).
Table 4.2.2 CDR enrichment by CDR1,2,3 nucleotide sequence (light chain)
| Count | CDR1nt | CDR1aa | CDR2nt | CDR2aa | CDR3nt | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|---|---|---|---|---|
| 11289 | GGTGGGTCGTTTAGTGGTTTCGAC | GGSFSGFD | ATCAGCCACACTGGAACTACG | ISHTGTT | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV3-2*02 | IGHJ3*02 |
| 3100 | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATCAGGAGATCCGGACTCCTTAATGATGCTTTTGATATCTGG | CARLPIRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 2339 | GGATTCATCTTCAGCACCTACTGG | GFIFSTYW | ATTAATGAAGATGGCAGGATTACC | INEDGRIT | TGTGTTAGAAGGCACCCAGCACCAACTGGCAACATTTTTGACTTCTGG | CVRRHPAPTGNIFDFW | IGHV5-6-3*01 | IGHJ2*01 |
| 1980 | GGTGACTCCATCAGCAATACTAGATATTAC | GDSISNTRYY | ATATATAATAGTGGAAATATC | IYNSGNI | TGTGCGGGGCACGTTTGGAACTACGAAGTTGACTACTGG | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 1452 | GGTGTCGCCATCACTAGTTTCCAC | GVAITSFH | ATATATCACAATGGAGACACC | IYHNGDT | TGTGCGAGAGTTGATGCAATCATTGAAATGGACTACTTCTACGGTCTGGACGTCTGG | CARVDAIIEMDYFYGLDVW | IGHV3-2*02 | IGHJ1*01 |
| 1339 | GGTGACTCCATCAGCAATACTAGATATTAC | GDSISNTRYY | ATATATAATAGTAGAAATATC | IYNSRNI | TGTGCGGGGCACGTTTGGAACTACGAAGTTGACTACTGG | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 976 | GGGTTCTCACTCAATACTCGTGGAACGACT | GFSLNTRGTT | ATTTATTGGGATGATGATAGC | IYWDDDS | TGTGCACACGGACGCCCAGACTGGGGAGCAGATGCTTTTGATGTCTGG | CAHGRPDWGADAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 931 | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATCAGGAGATCCGGGCTCCATAATGATGCTTTTGATATATGG | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 829 | GGCGGCTCCATCAGTAGCAGTAGTTACCAC | GGSISSSSYH | ATCTATTATAGTGGGAGCACG | IYYSGST | TGTGCGAGTCGTCGAAATGAACCTGGAGGGTGGTTCGACTCCTGG | CASRRNEPGGWFDSW | IGHV3-1*01 | IGHJ3*01 |
| 753 | GGTGGGTCATTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTATC | ISHSGSI | TGTGCGAGACTTCCCATGAGGAGATCCGGGCTCCTTAATGATGCTTTTGATATCTGG | CARLPMRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 705 | GGTTACACCTTTACTGATTATGCT | GYTFTDYA | ATCAGCGTTTCCAATGGTAAAACA | ISVSNGKT | TGTGCGAGAGCGTTTCAACCTCAAGTCTGGGTCGGGGAGTCTTATCTCGACTACTGG | CARAFQPQVWVGESYLDYW | IGHV1-77*01 | IGHJ2*01 |
| 666 | GGTGGGTCCTTTAGTGGTTACGAC | GGSFSGYD | ATCAGTCACAGTGGAAGTACC | ISHSGST | TGTGCGAGACTTCCCATCAGGAGATCCGGGCTCCATAATGATGCTTTTGATATATGG | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 626 | NNNNNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXXX | NNNNNNNNNNNNNNNNNNNNN | XXXXXXX | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV12-3*01 | IGHJ3*02 |
| 611 | GGGTTCTCATTCACTTCTAGTGGACGGGGT | GFSFTSSGRG | ATTTATTGGGATGACGATAAG | IYWDDDK | TGTGCACACAGACCACCATATCAAGGGTACTACTACTTTGACTATTGG | CAHRPPYQGYYYFDYW | IGHV8-12*01 | IGHJ2*01 |
| 602 | GGGTTCTCATTCAATACTCTTGGAACGACT | GFSFNTLGTT | ATTTATTGGGATGATGATAGC | IYWDDDS | TGTGCACACGGACGCCCAGACTGGGGAGAAGATGCTTTTGATGTCTGG | CAHGRPDWGEDAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 572 | GGCTTCTCATTCAGTTCTAGTGGACTGGGT | GFSFSSSGLG | ATTTATTGGGATGATGATAAG | IYWDDDK | TGTGCACACAGACCGCCATATCAACGGTATTACTACTTTGACTATTGG | CAHRPPYQRYYYFDYW | IGHV8-6*01 | IGHJ2*01 |
| 514 | GGATACACGTTCACCAATTATGCT | GYTFTNYA | ATGGACCCGAATAGCGGCGACACA | MDPNSGDT | TGTGCGAGGACCAACTGGGCAGCCTACGGTGTCCCCGACTACTGG | CARTNWAAYGVPDYW | IGHV1-72*04 | IGHJ4*01 |
| 485 | NNNNNNNNNNNNNNNNNNNNNNNNNNN | XXXXXXXXX | NNNNNNNNNNNNNNNNNNNNN | XXXXXXX | TGTGCGCGAATTCCCATGAGGAGAACCGGGGTCAACGATGATGCCTTTGATATGTGG | CARIPMRRTGVNDDAFDMW | IGHV12-3*02 | IGHJ3*02 |
| 459 | GGTGACTCCGTCAGCAATCATAAATACTAC | GDSVSNHKYY | ATCTATTCTGGTGGGAACACC | IYSGGNT | TGTGCGGGGCACAATTGGAATTACGAGGTTGACTACTGG | CAGHNWNYEVDYW | IGHV3-5*02 | IGHJ4*01 |
| 455 | GGATTCTCACTCACTACTCGTGGAGGGGGT | GFSLTTRGGG | CTTTATTGGGATGAAAAGACA | LYWDEKT | TGTGCACACCGTATTGGTTCGGAGACATACTTCGACTACTGG | CAHRIGSETYFDYW | IGHV8-12*01 | IGHJ2*01 |
Shown is the partial result of one sample (ASC-S-vH).
If the reads collapsed on nucleotide sequence did not have replicates, they were deleted from subsequent analysis for the sake of sequence accuracy and result reliability.
Table 4.2.3 CDR enrichment by CDR amino acid sequence (heavy chain)
| Count | CDR1aa | CDR2aa | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|---|---|
| 11412 | GGSFSGFD | ISHTGTT | CARIPMRRTGVNDDAFDMW | IGHV3-2*02 | IGHJ3*02 |
| 3317 | GGSFSGYD | ISHSGSI | CARLPIRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 2444 | GFIFSTYW | INEDGRIT | CVRRHPAPTGNIFDFW | IGHV5-6-3*01 | IGHJ2*01 |
| 2037 | GDSISNTRYY | IYNSGNI | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 1634 | GFSLNTRGTT | IYWDDDS | CAHGRPDWGADAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 1472 | GVAITSFH | IYHNGDT | CARVDAIIEMDYFYGLDVW | IGHV3-2*02 | IGHJ1*01 |
| 1347 | GDSISNTRYY | IYNSRNI | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 1169 | GGSFSGYD | ISHSGSI | CARLPMRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 965 | GGSFSGYD | ISHSGSI | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 836 | GGSISSSSYH | IYYSGST | CASRRNEPGGWFDSW | IGHV3-1*01 | IGHJ3*01 |
| 763 | GYTFTDYA | ISVSNGKT | CARAFQPQVWVGESYLDYW | IGHV1-77*01 | IGHJ2*01 |
| 692 | GGSFSGYD | ISHSGST | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 637 | XXXXXXXXX | XXXXXXX | CARIPMRRTGVNDDAFDMW | IGHV12-3*01 | IGHJ3*02 |
| 616 | GFSFTSSGRG | IYWDDDK | CAHRPPYQGYYYFDYW | IGHV8-12*01 | IGHJ2*01 |
| 609 | GFSLSTTGVG | IYWDDDR | CARQNSGYDWNSRCYDYW | IGHV8-12*01 | IGHJ4*01 |
| 604 | GFSFNTLGTT | IYWDDDS | CAHGRPDWGEDAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 577 | GFSFSSSGLG | IYWDDDK | CAHRPPYQRYYYFDYW | IGHV8-6*01 | IGHJ2*01 |
| 519 | GYTFTNYA | MDPNSGDT | CARTNWAAYGVPDYW | IGHV1-72*04 | IGHJ4*01 |
| 509 | GFSLNTRGTT | IYWDGDD | CAHGRPDWGADAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 498 | XXXXXXXXX | XXXXXXX | CARIPMRRTGVNDDAFDMW | IGHV12-3*02 | IGHJ3*02 |
Shown is the partial result of one sample (ASC-S-vH).
Table 4.2.4 CDR enrichment by CDR amino acid sequence (light chain)
| Count | CDR1aa | CDR2aa | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|---|---|
| 7248 | QGISRG | YAS | CQQFNQYPITF | IGKV11-125*01 | IGKJ4*01 |
| 4116 | QGIGSY | AAS | CQQLSSYPLTF | IGKV11-125*01 | IGKJ1*01 |
| 3135 | QGIGNE | AAS | CLQHKNHVWTF | IGKV11-125*01 | IGKJ1*01 |
| 2934 | QSISSG | DAS | CQQYNNYPITF | IGKV11-125*01 | IGKJ4*01 |
| 1919 | QAIGGG | YAS | CQQFNAYPITF | IGKV11-125*01 | IGKJ4*01 |
| 1473 | QSFGCC | GAS | CHQRSGWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| 1374 | QSISSG | DAS | CQQYNSYPITF | IGKV11-125*01 | IGKJ4*01 |
| 1373 | QSFGCC | GAS | CHQRSDWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| 1280 | QSVSSN | DAS | CQQYNNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
| 804 | QSVLHSSNNKNY | WSS | CQQHYIIPWTF | IGKV8-30*01 | IGKJ1*01 |
| 788 | QSISSG | NAS | CQQYNVYPITF | IGKV11-125*01 | IGKJ4*01 |
| 735 | QSVSSN | GAS | CQQYNNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
| 593 | QGINTA | DAS | CQHFNSFPLAF | IGKV11-125*01 | IGKJ1*01 |
| 579 | QSVSSNY | GAS | CQQYGTSPWTF | IGKV18-36*01 | IGKJ1*01 |
| 578 | ESVSSY | DAS | CQQRSGWPWTF | IGKV18-36*01 | IGKJ1*01 |
| 539 | QSFGCC | GAS | CHQRSSWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| 535 | QDIGSS | GAS | CQQLKSYPINF | IGKV11-125*01 | IGKJ1*01 |
| 449 | RGVGSN | GAS | CQQYDDWPPWTF | IGKV18-36*01 | IGKJ1*01 |
| 448 | QSISSG | DAS | CQQYNAYPITF | IGKV11-125*01 | IGKJ4*01 |
| 419 | QSVSSN | GAS | CQQYDNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
Shown is the partial result of one sample (ASC-S-vL).
Table 4.2.5 CDR enrichment by CDR3 amino acid sequence (heavy chain)
| Count | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|
| 11978 | CARIPMRRTGVNDDAFDMW | IGHV3-2*02 | IGHJ3*02 |
| 3733 | CARLPIRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 3662 | CAGHVWNYEVDYW | IGHV3-5*02 | IGHJ2*01 |
| 2971 | CVRRHPAPTGNIFDFW | IGHV5-6-3*01 | IGHJ2*01 |
| 2778 | CAHGRPDWGADAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 2213 | CAHGRPDWGEDAFDVW | IGHV8-12*01 | IGHJ1*01 |
| 1877 | CARLPIRRSGLHNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 1842 | CARVDAIIEMDYFYGLDVW | IGHV3-2*02 | IGHJ1*01 |
| 1527 | CAGHNWNYEVDYW | IGHV3-5*02 | IGHJ4*01 |
| 1403 | CARDGKRTYSYDRGEDYW | IGHV5-17*01 | IGHJ4*01 |
| 1298 | CARLPMRRSGLLNDAFDIW | IGHV3-2*02 | IGHJ3*01 |
| 1228 | CARIPMRRTGVNDDAFDMW | IGHV12-3*02 | IGHJ3*02 |
| 950 | CARQNSGYDWNSRCYDYW | IGHV8-12*01 | IGHJ4*01 |
| 855 | CASRRNEPGGWFDSW | IGHV3-1*01 | IGHJ3*01 |
| 827 | CARAFQPQVWVGESYLDYW | IGHV1-77*01 | IGHJ2*01 |
| 655 | CARCRGDSNYGWYDPW | IGHV3-5*02 | IGHJ3*01 |
| 645 | CARIPMRRTGVNDDAFDMW | IGHV12-3*01 | IGHJ3*02 |
| 638 | CAHRPPYQGYYYFDYW | IGHV8-12*01 | IGHJ2*01 |
| 603 | CAHRPPYQRYYYFDYW | IGHV8-6*01 | IGHJ2*01 |
| 577 | CATGPTMVMLDYW | IGHV1-64*01 | IGHJ2*01 |
Shown is the partial result of one sample (ASC-S-vH).
Table 4.2.6 CDR enrichment by CDR3 amino acid sequence (light chain)
| Count | CDR3aa | Top_V_Gene | Top_J_Gene |
|---|---|---|---|
| 8864 | CQQFNQYPITF | IGKV11-125*01 | IGKJ4*01 |
| 5516 | CQQLSSYPLTF | IGKV11-125*01 | IGKJ1*01 |
| 4171 | CQQYNNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
| 3892 | CLQHKNHVWTF | IGKV11-125*01 | IGKJ1*01 |
| 3741 | CQQYNNYPITF | IGKV11-125*01 | IGKJ4*01 |
| 2536 | CQQFNAYPITF | IGKV11-125*01 | IGKJ4*01 |
| 2162 | CHQRSDWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| 2079 | CHQRSGWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| 1744 | CQQHYIIPWTF | IGKV8-30*01 | IGKJ1*01 |
| 1737 | CQQRSGWPWTF | IGKV18-36*01 | IGKJ1*01 |
| 1697 | CQQYNSYPITF | IGKV11-125*01 | IGKJ4*01 |
| 1668 | CLQHYNHVWTF | IGKV11-125*01 | IGKJ1*01 |
| 1022 | CQQYNVYPITF | IGKV11-125*01 | IGKJ4*01 |
| 996 | CQQYGTSPWTF | IGKV18-36*01 | IGKJ1*01 |
| 987 | CQQYNNWPLWTF | IGKV18-36*01 | IGKJ1*01 |
| 780 | CHQRSSWPPFTF | IGKV18-36*01 | IGKJ4*01 |
| 722 | CQQLKSYPINF | IGKV11-125*01 | IGKJ1*01 |
| 690 | CQQYDNWPLWTF | IGKV5-45*01 | IGKJ1*01 |
| 686 | CQHFNSFPLAF | IGKV11-125*01 | IGKJ1*01 |
| 683 | CQQYNNYWTF | IGKV18-36*01 | IGKJ1*01 |
Shown is the partial result of one sample (ASC-S-vL).
CDR3 amino acid sequences were further extracted and the length distribution was plotted as in Figure 4.3.
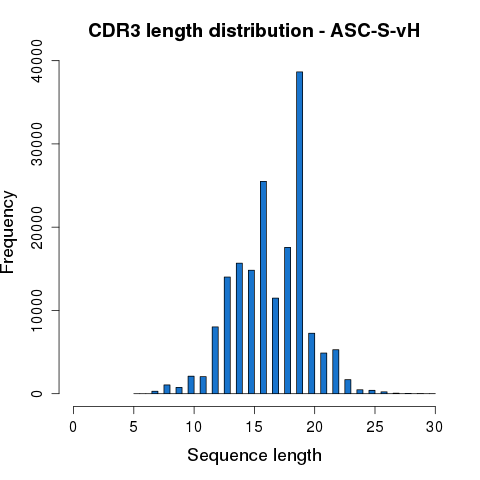
Figure 4.3 CDR3 amino acid sequence length distribution.
V(D)J usage of sequences were analyzed. Sequences were collapsed on V(D)J combination. The proportions of the top ten combinations in the heavy and light chain populations are shown in Figure 4.4.1 and Figure 4.4.2, respectively.
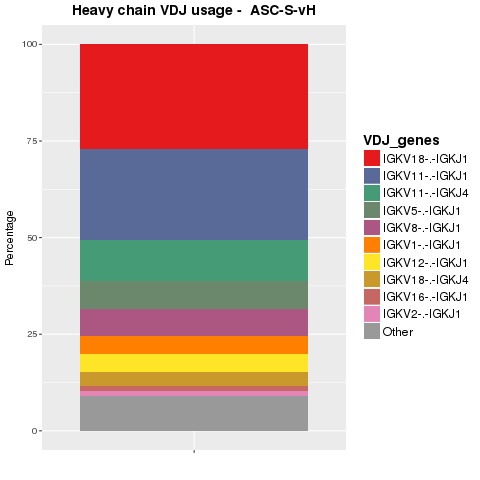
Figure 4.4.1 Ig top VDJ combination usage for heavy chains.
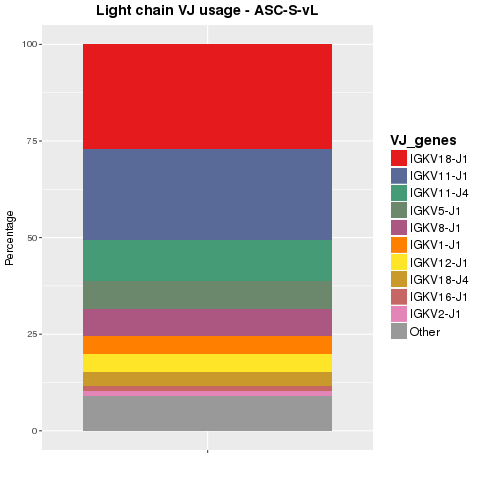
Figure 4.4.2 Ig top VJ combination usage for light chains.
To assess germline gene usage, V gene annotation information was extracted for all the sequences and the total count of each germline gene was calculated and summarized in Tables 4.5.1 and 4.5.2.
Table 4.5.1 Heavy chain V gene usage
| Top_V_Gene | ASC-S-vH | PBMC-0-S-vH | PBMC-7-S-vH |
|---|---|---|---|
| IGHV1 | 26385 | 33920 | 28208 |
| IGHV10 | 1418 | 1868 | 1583 |
| IGHV10S3 | 16 | 10 | 7 |
| IGHV10S4 | 1 | NA | 1 |
| IGHV12 | 3858 | 2548 | 3551 |
| IGHV13 | 31 | 4 | 20 |
| IGHV14 | 919 | 1738 | 1189 |
| IGHV14S4 | 2 | NA | NA |
| IGHV15 | 21 | NA | NA |
| IGHV16 | 612 | 386 | 406 |
| IGHV1S12 | 577 | 258 | 322 |
| IGHV1S126 | 647 | 523 | 317 |
| IGHV1S127 | 510 | 436 | 250 |
| IGHV1S130 | 3 | 4 | NA |
| IGHV1S132 | 10 | 15 | 4 |
| IGHV1S134 | 38 | 15 | 68 |
| IGHV1S136 | 4 | 13 | 28 |
| IGHV1S137 | 1 | 1 | 9 |
| IGHV1S14 | 2 | NA | 1 |
| IGHV1S15 | 51 | 18 | 109 |
| IGHV1S17 | 1 | 1 | 15 |
| IGHV1S18 | 7 | 258 | 141 |
| IGHV1S19 | 4 | 12 | 103 |
| IGHV1S22 | 20 | 19 | 2 |
| IGHV1S26 | 697 | 270 | 322 |
| IGHV1S28 | 21 | 21 | 6 |
| IGHV1S29 | 14 | 26 | 6 |
| IGHV1S34 | 4 | 14 | 12 |
| IGHV1S45 | 190 | 25 | 120 |
| IGHV1S46 | 408 | 1032 | 600 |
| IGHV1S5 | 2 | 7 | 2 |
| IGHV1S50 | 31 | 83 | 58 |
| IGHV1S55 | 7 | 2 | 6 |
| IGHV1S56 | 178 | 146 | 635 |
| IGHV1S61 | 48 | 53 | 41 |
| IGHV1S81 | 111 | 55 | 24 |
| IGHV2 | 42 | 21 | 21 |
| IGHV3 | 61675 | 40187 | 56460 |
| IGHV3S1 | 8800 | 21984 | 15880 |
| IGHV3S7 | 16 | 2 | 12 |
| IGHV4 | 22 | 1 | 14 |
| IGHV5 | 35995 | 35220 | 36499 |
| IGHV5S12 | 31 | 77 | 103 |
| IGHV5S21 | 266 | 166 | 113 |
| IGHV5S24 | 18 | 1 | 11 |
| IGHV5S4 | 59 | 9 | 44 |
| IGHV5S9 | 1 | 2 | 8 |
| IGHV6 | 455 | 333 | 428 |
| IGHV7 | 1481 | 1333 | 1406 |
| IGHV8 | 26539 | 30520 | 22482 |
| IGHV8S2 | 28 | 9 | 6 |
| IGHV9 | 36 | 37 | 44 |
| IGHV1S16 | NA | 1 | NA |
| IGHV1S47 | NA | 1 | NA |
| IGHV2S3 | NA | 1 | NA |
Table 4.5.2 Light chain V gene usage
| Top_V_Gene | ASC-S-vL | PBMC-0-S-vL | PBMC-7-S-vL |
|---|---|---|---|
| IGKV1 | 7420 | 4924 | 5979 |
| IGKV10 | 506 | 309 | 351 |
| IGKV11 | 49516 | 36390 | 41839 |
| IGKV12 | 4256 | 7743 | 5958 |
| IGKV13 | 115 | 53 | 85 |
| IGKV14 | 327 | 128 | 245 |
| IGKV15 | 1261 | 1100 | 1443 |
| IGKV16 | 3775 | 2143 | 1970 |
| IGKV17 | 2 | 3 | 2 |
| IGKV18 | 31877 | 42729 | 38078 |
| IGKV19 | 127 | 43 | 24 |
| IGKV2 | 1159 | 1714 | 1566 |
| IGKV3 | 62 | 37 | 57 |
| IGKV4 | 100 | 18 | 32 |
| IGKV5 | 10033 | 8702 | 8996 |
| IGKV6 | 483 | 744 | 597 |
| IGKV8 | 8066 | 8131 | 8794 |
| IGKV9 | 85 | 27 | 34 |
| IGLV1 | NA | 2 | NA |
The distribution of V gene usage is also illustrated in bar graphs (Figure 4.5.1 and Figure 4.5.2).
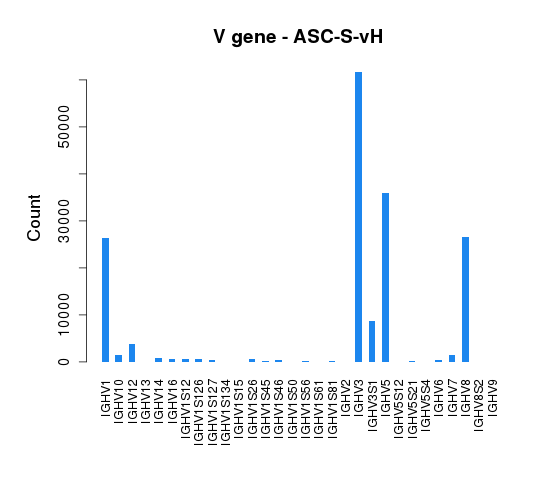
Figure 4.5.1 V gene usage of heavy chains.
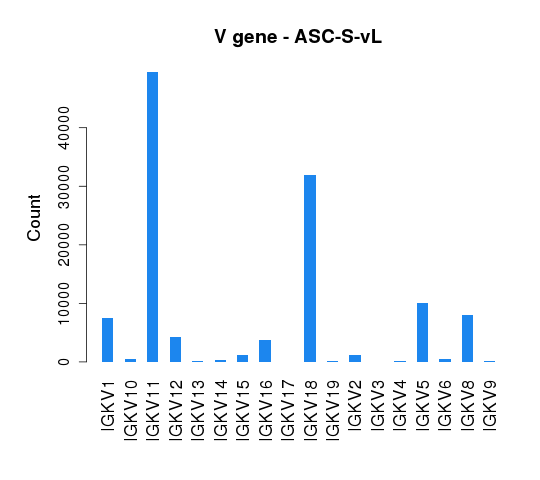
Figure 4.5.2 V gene usage of light chains.
To analyze CDR3 amino acid usage frequency, CDR3 sequences were clustered according to similarity (threshold: 0.8). The summary of top 20 clusters of each sample is as in Table 4.6. The complete results for all the samples are in the 'Report\Clustering_summary_tables' folder and can be accessed using the link below:
The logo of the most representative sequence of each cluster [6] is in the 'Report\Clustering_weblogo' folder and can be viewed using the links below:
Table 4.6 Top 20 CDR3 clusters and analysis summary
| Cluster_ID | Total_cnt | Freq_total_cnt | Unique_cnt | Freq_unique_cnt | CDR3_seq |
|---|---|---|---|---|---|
| Cluster_0 | 14678 | 8.5182 | 89 | 1.3035 | 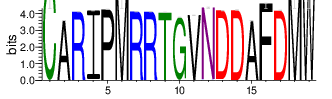
|
| Cluster_1 | 10147 | 5.8887 | 126 | 1.8453 | 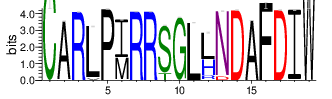
|
| Cluster_2 | 6939 | 4.027 | 65 | 0.952 | 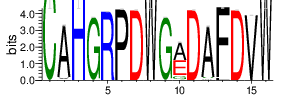
|
| Cluster_3 | 6641 | 3.854 | 57 | 0.8348 | 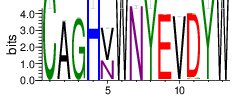
|
| Cluster_4 | 3964 | 2.3005 | 28 | 0.4101 | 
|
| Cluster_5 | 3318 | 1.9256 | 58 | 0.8494 | 
|
| Cluster_6 | 2574 | 1.4938 | 35 | 0.5126 | 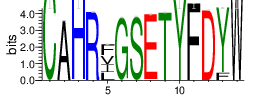
|
| Cluster_7 | 2007 | 1.1647 | 28 | 0.4101 | 
|
| Cluster_8 | 1737 | 1.008 | 26 | 0.3808 | 
|
Shown is the partial result of one sample (ASC-S-vH). Cluster_ID: the identification number of the cluster; Total_cnt: the total clonal count of CDR3 sequences in the cluster; Freq_total_cnt: the sequence count percentage of the entire read count of the sample; Unique_cnt: the count of unique CDR3 sequences in the cluster; Freq_total_cnt: the unique sequence count percentage of the total unique count of the sample; CDR3_seq: the most abundant CDR3 amino acid sequence; CDR3_length: the length of the most abundant CDR3 amino acid sequence; weblogo of the representative sequence of the cluster can be found in ‘Top20_clusters/weblogo’ directory.
The clonal abundance distribution was calculated with confidence intervals derived via bootstrapping. The clonal diversity of the repertoire was accessed using diversity index [7]. Abundance curves are in figure 4.7.1 and diversity curves are in Figure 4.7.2.
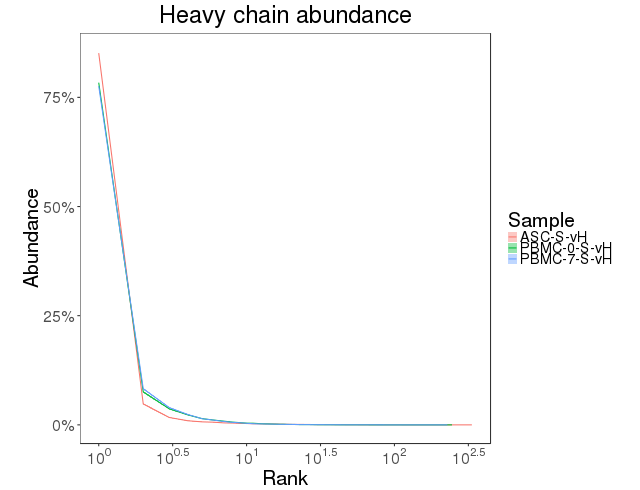
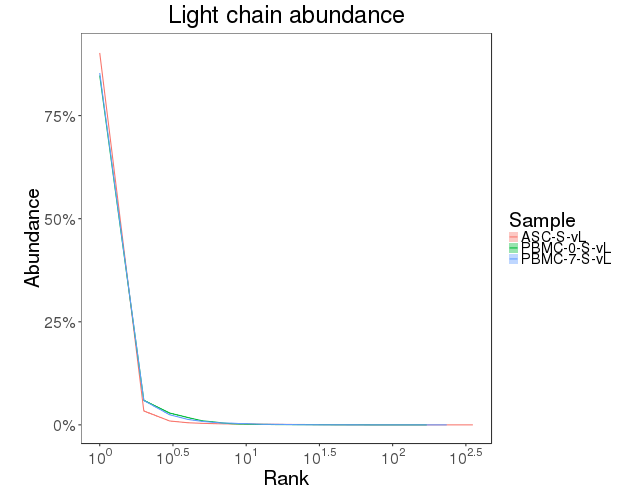
Figure 4.7.1 Abundance curves.
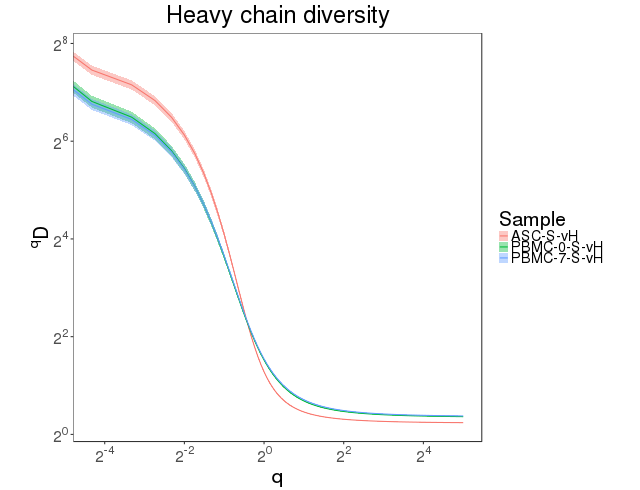
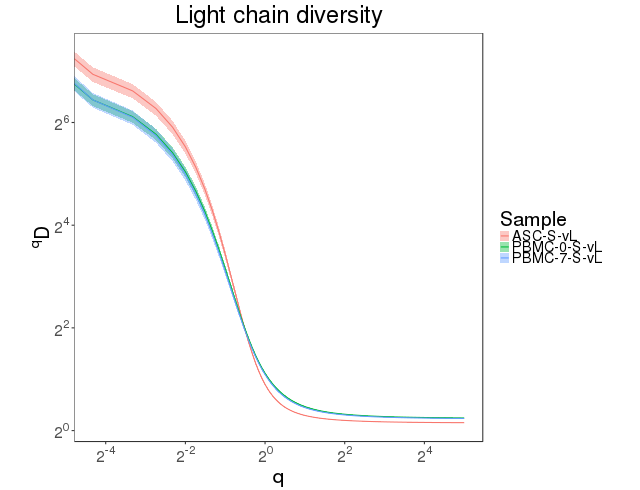
Figure 4.7.2 Diversity curves.
[1] Murphy K. Janeway's Immunobiology. New York: Garland Science (2012).
[2] Lefranc, MP. et al., The international ImMunoGeneTics database. Nucleic Acids Res. (1999) 27 (1): 209-212.
[3] Bolger, AM. et al., Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30(15): 2114–2120.
[4] Andrews S. et al., FastQC: a quality control tool for high throughput sequence data. (2010) Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
[5] Ye J. et al., IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. (2013) 41: W34–W40.
[6] Crooks GE. et al., WebLogo: a sequence logo generator. Genome Res. (2004) 14(6):1188-90.
[7] Hill, M. et al., Diversity and evenness: a unifying notation and its consequences. Ecology (1973) 54:427-432.